Launching the GUI
Launch the graphical interface by double-clicking the SynapSpec application icon:
- Linux: Double-click
SynapSpecin Applications folder - macOS: Double-click
SynapSpecin Applications folder - Windows: Double-click
SynapSpec.exein the installation folder
Interface Overview
Main Window Layout
The GUI is organized with a tab-based interface:
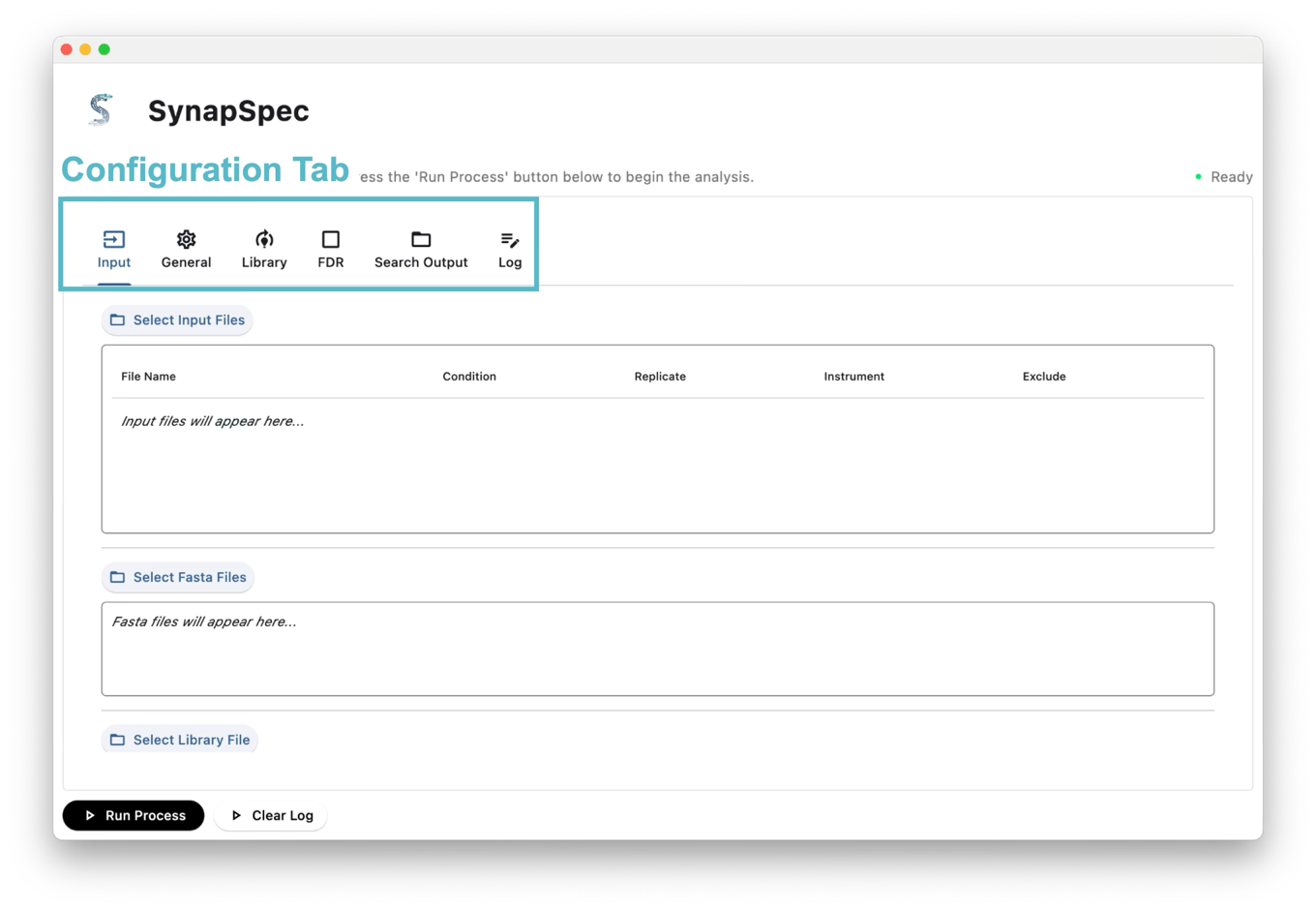
Tab Structure
- Input: Configure input files (MS data, FASTA/Library) and output directory
- General: Basic configuration options
- Library: Library specific settings
- FDR: False Discovery Rate parameters
- Search Output: Output report related settings
- Log: View real-time processing logs
Control Elements
- Run Process Button: Located at the bottom to start analysis
- Status Indicator: Located in the upper right corner, displays current process status
Loading Data
Supported File Types
- Raw Files: .raw, .mzML
- Fasta Files: .fasta
- Library Files: .tsv, .txt
- Output formats: .csv, .tsv, .parquet
Configuring Input Settings
Navigate to the Input tab and configure the following:
Step 1: Input Files - Click Select Input Files to choose your mass spectrometry data (.raw or .mzML)
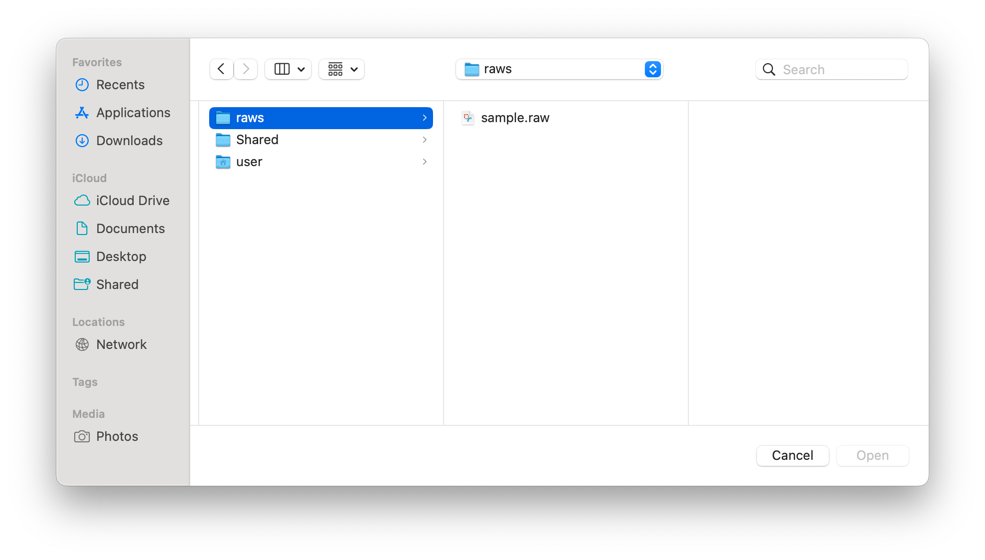
Step 2: FASTA or Library File (choose one)
- Click
Select Fasta Filesfor protein sequence database search, OR - Click
Select Library Filefor spectral library search (.tsv or .txt)
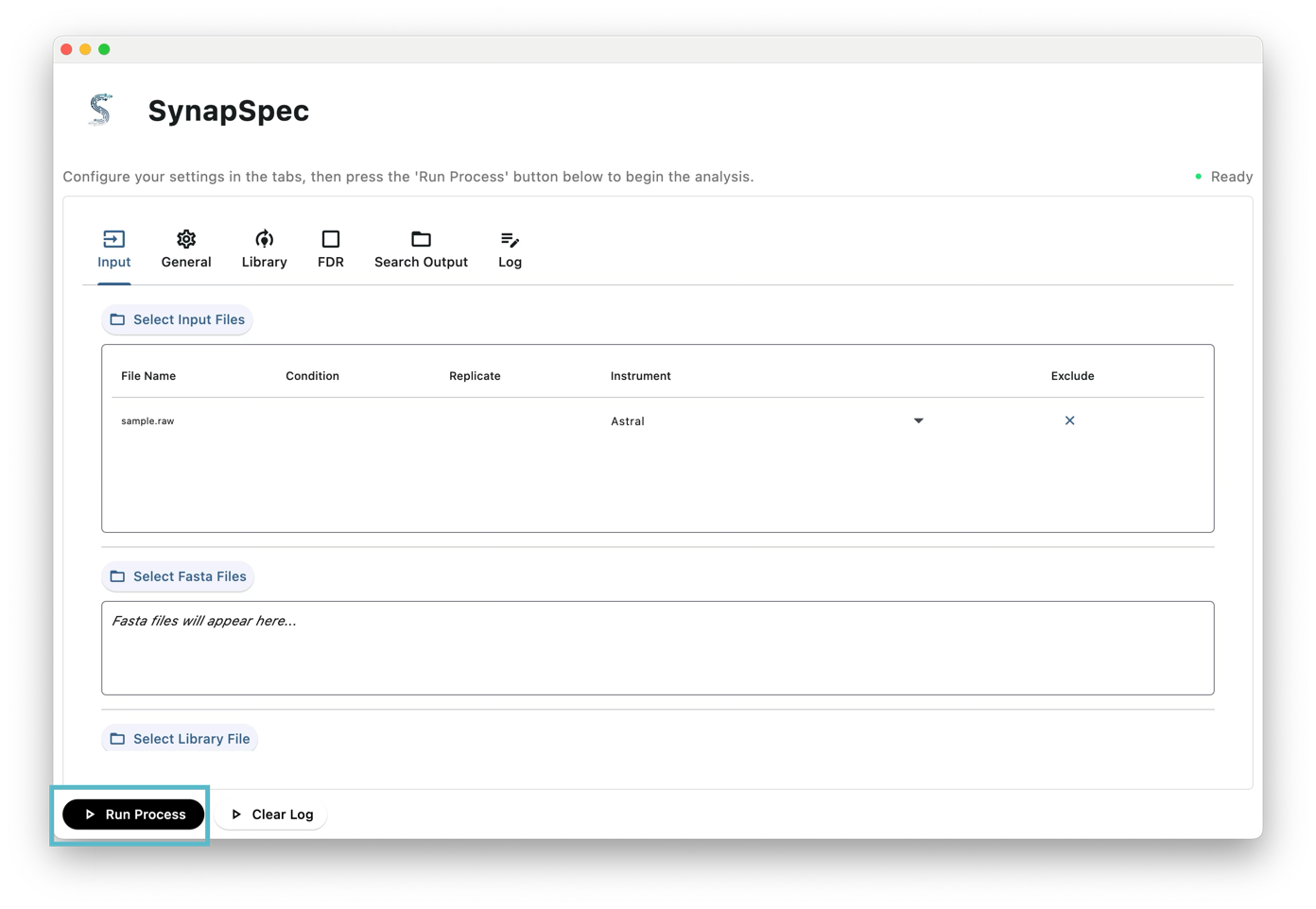
Step 3: Output Directory - Click Select Output Directory to choose where results will be saved
Step 4: Click “Run Process”
Output Results
After the analysis completes, SynapSpec generates the following output files in your specified output directory